Instructions of using HIVPRE
HIVPRE is constructed based on the Genome-build GRCh38.p12.
It provides high-throughput predictions for putative HIV-1 interacting proteins and their directionalities.
You can quickly make your own predictions by following a few steps.
For more information please contact us via: david.l.robertson@glasgow.ac.uk or h.chai.research@gmail.com
STEP ONE
You do not need to input or upload your genetic or proteomic sequences. The only thing HIVPRE wants is the identifier of your proteins.
HIVPRE accepts SIX types of gene or protein identifiers including: HGNC ID, RefSeqAcc, Transcipt ID, Approved Symbol, NCBI gene ID and Ensembl gene ID.
You can input different types of identifiers or unify them with the matching service from UniProt.
HIVPRE does not set restrictions on the number of your identifiers, but they should be valid in the right format and be separated by commas or newlines:

In this example, Q1 is a gene identifier used by HUGO Gene Nomenclature Committee. Please include 'HGNC:' if you would like to use such identifiers.
Q5 is a gene identifier used by National Center for Biotechnology Information. As a matter of priority, numerical identifiers will be matched to NCBI IDs.
Q6 & Q3 are gene and transcript identifiers used by Ensembl. You may find multiple results for a single human gene with more than one open reading frames.
Q2 is a protein identifier used by NCBI Reference Sequence Database.
Q4 is an approved symbol for a specific gene or protein. HIVPRE provides case sensitive matching for approved symbols. Thus do not input symbols like Q8.
Please note, if you want to make predictions with your HGNC ID but forget to include 'HGNC:', exemplified by Q7, HIVPRE will recognise it as a NCBI ID.
STEP TWO
You can remove all you inputs by click the CLEAR button or get the results of your prediction soon after you click the PREDICT button on the right:
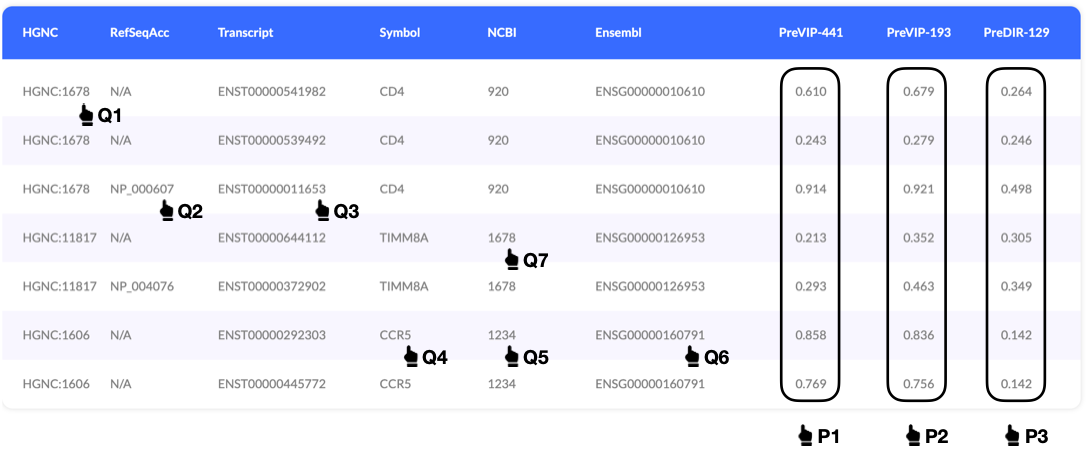
In this example, we get 7 prediction results based on the input 8 identifiers.
You will find your identifiers in the former six columns and prediction results generated by three models in the later three columns.
Gene identifier Q1 gets three prediction results as CD4 gene three available open reading frames.
Identifier Q7 is matched to TIMM8A rather than CD4 for not including 'HGNC:' before '1678'.
P1 are predicted by model PreVIP-441, which is designed with 441 features for the prediction of putative human proteins interacting with HIV-1 proteins.
The prediction score can be used to measure the probability of being a HIV-1 interacting protein (VIPs) for your protein of interest.
In general, higher score indicates higher possibility of being a VIP. But in specific cases, it need to be treated with caution.
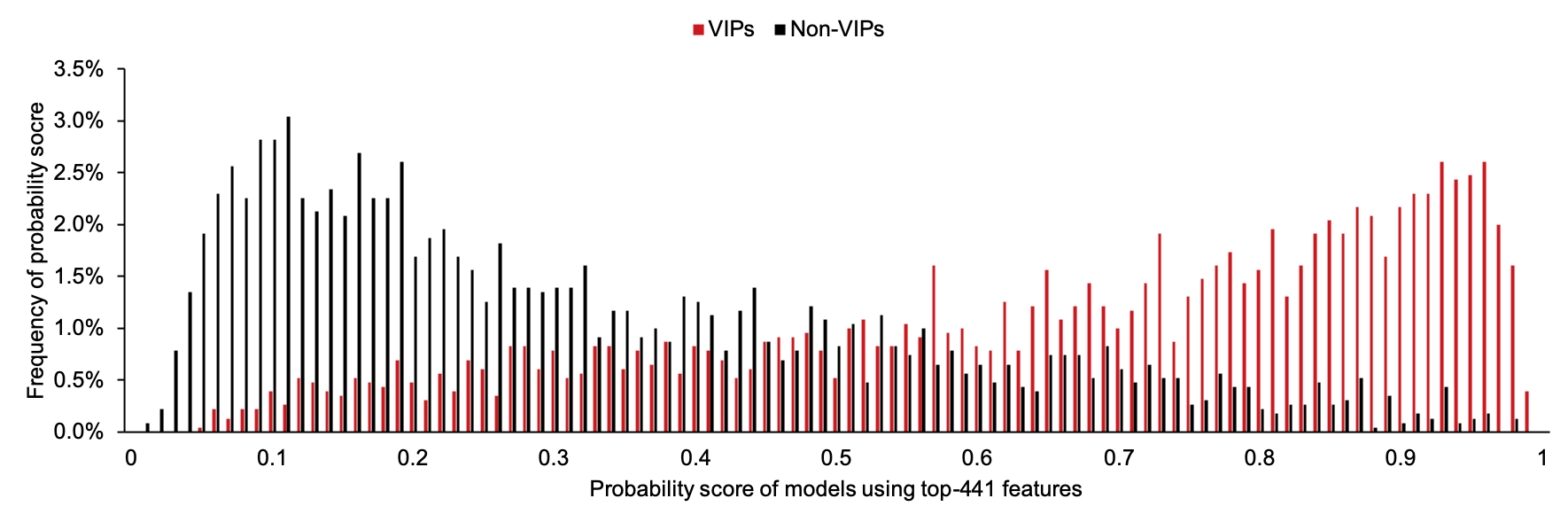
We recommend to use 0.73 as the threshold to judge results generated by model PreVIP-441.
Here is the distribution of prediction scores generated by model with 441 features for VIPs and non-VIPs on our balanced training dataset S1':
P2 are predicted by model PreVIP-193, which is designed with 193 features also for the prediction of putative human proteins interacting with HIV-1 proteins.
In general, higher score indicates higher possibility of being a VIP. But in specific cases, it need to be treated with caution.
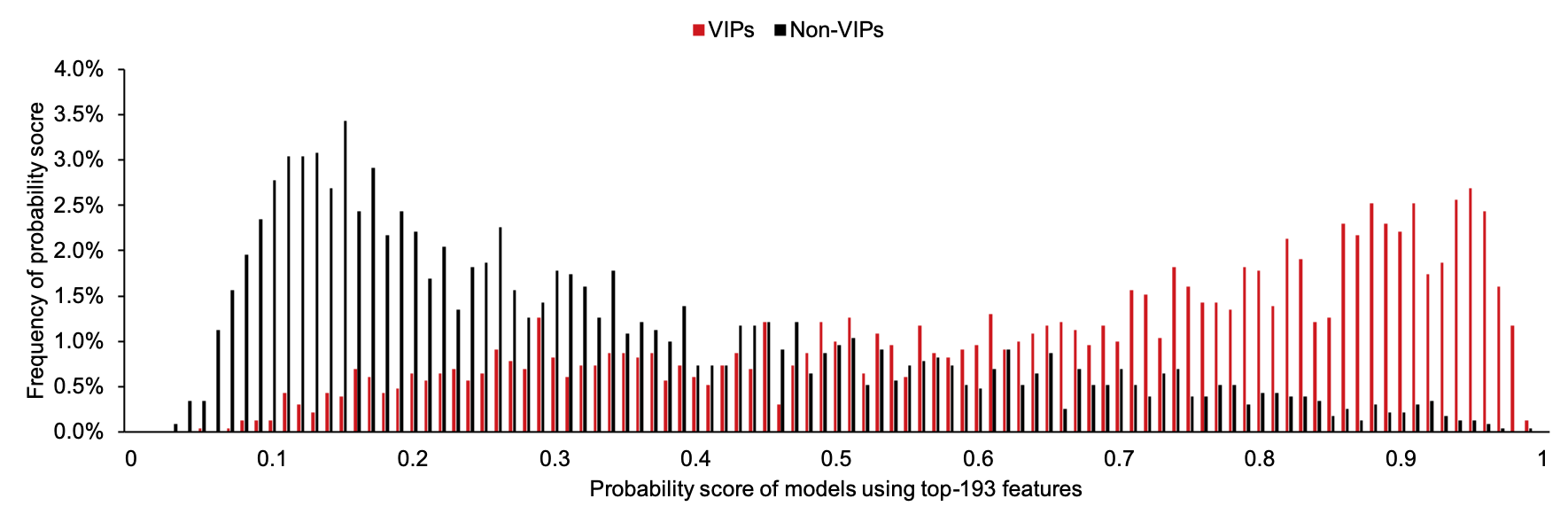
We recommend to use 0.82 as the threshold to judge results generated by model PreVIP-193.
Here is the distribution of prediction scores generated by model with 193 features for VIPs and non-VIPs on our balanced training dataset S1':
P3 are predicted by model PreDIR-129, which is designed with 129 features for the prediction of directionality of putative HIV-1-host protein-protein interactions.
Here is the distribution of prediction scores generated by model with 129 features for pro-host (backward), pro-viral (forward) and bidirectional VIPs on our datasets:
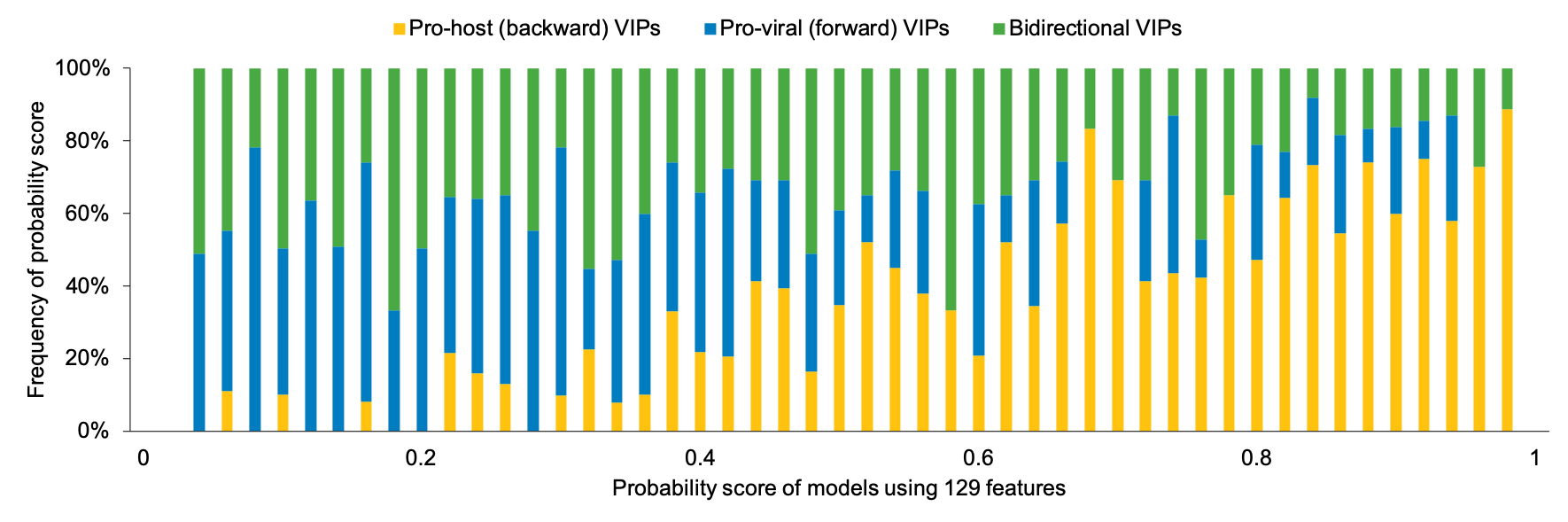
Proteins with higher prediction scores are more likely to be 'backward' VIPs but clues for 'forward' and 'bidirectional' VIPs are not so clear.
We figure that ‘forward’ VIPs might potentially respond to HIV-1 infection and target HIV-1, making them ‘bidirectional’.
Please check our supplementary file for more detailed recommendation about the prediction score generated by PreDIR-129.